


ABOUT THE PROJECT
During the drug screening process, thousands of compounds are tested at a time in search for specific compounds that can be used to treat illnesses. These compounds go through numerous bioassay tests, and from the thousands of compounds tested, only a few will actually be active. Problems arise when the active compound produces a harmful side effect, and the way to determine if the active compound has side effects is to search for all the relevant bioassays, which have tested that specific molecule in a database. This task is often overwhelming, because current chemical and bioassay databases will “dump” information into a window. Sometimes that information is relevant, while other times users must sift through a large amount of information manually to find what they are looking for. The aim of this project is to develop a database that is capable of creating a profile for a compound of interest. Users can search for a compound and a list of all the relevant bioassays will appear. This information can potentially provide great details about the efficacy of the compound. This database will ultimately save researchers a great deal of time, save valuable resources, and promote advancements in drug discovery.
FEATURES
- MySQL
- JavaScript
- PHP
- HTML 5
- Navicat TM
- phpMyAdmin
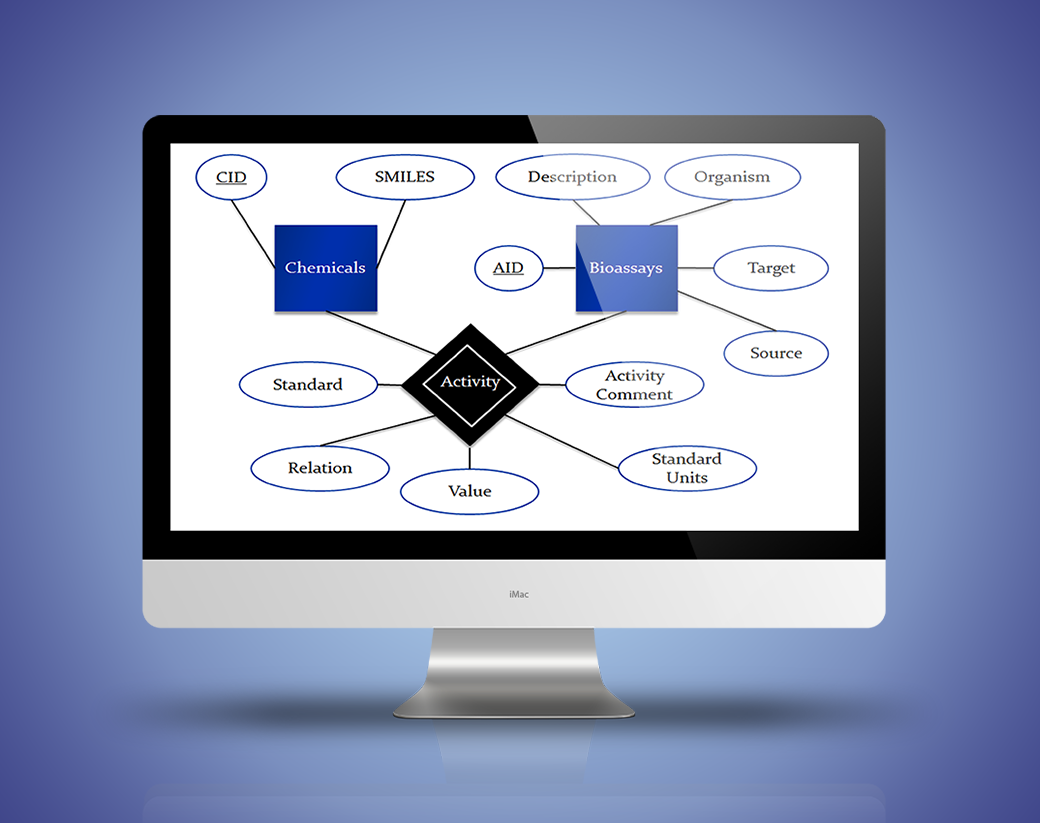
ENTITY-RELATIONSHIP (E/R)
- Chemicals(CID, SMILES)
- Bioassays(AID, Description, Organism, Target, Source)
- Activity(CID, AID, Standard, Relation, Value, Standard Units, Activity Comment)
FUNCTIONAL DEPENDENCIES
- In Chemicals: CID → SMILES
- In Bioassays: AID → Organism, Target, Source
- In Activity: CID, AID → Standard, Relation, Value, Standard Units
MADE FOR
Rutgers University